8. Phantom Data
DRSuite also provides a tool to generate synthetic multicontrast phantom data with from a known ground-truth spectroscopic image, and accommodates custom diffusion-relaxation encoding schemes. This can be useful for evaluating and comparing the performance of different acquisition protocols and spectrum estimation methods.
The numerical phantom is designed to possess interesting spectroscopic image features, and was obtained by simplifying the features observed in the mouse spinal cord injury datasets reported in the following paper (which should be cited, along with DRSuite, when using this phantom):
Kim, E. K. Doyle, J. L. Wisnowski, J. H. Kim, J. P. Haldar. Diffusion-Relaxation Correlation Spectroscopic Imaging: A Multidimensional Approach for Probing Microstructure. Magnetic Resonance in Medicine 78:2236-2249, 2017. <https://dx.doi.org/10.1002/mrm.26629>
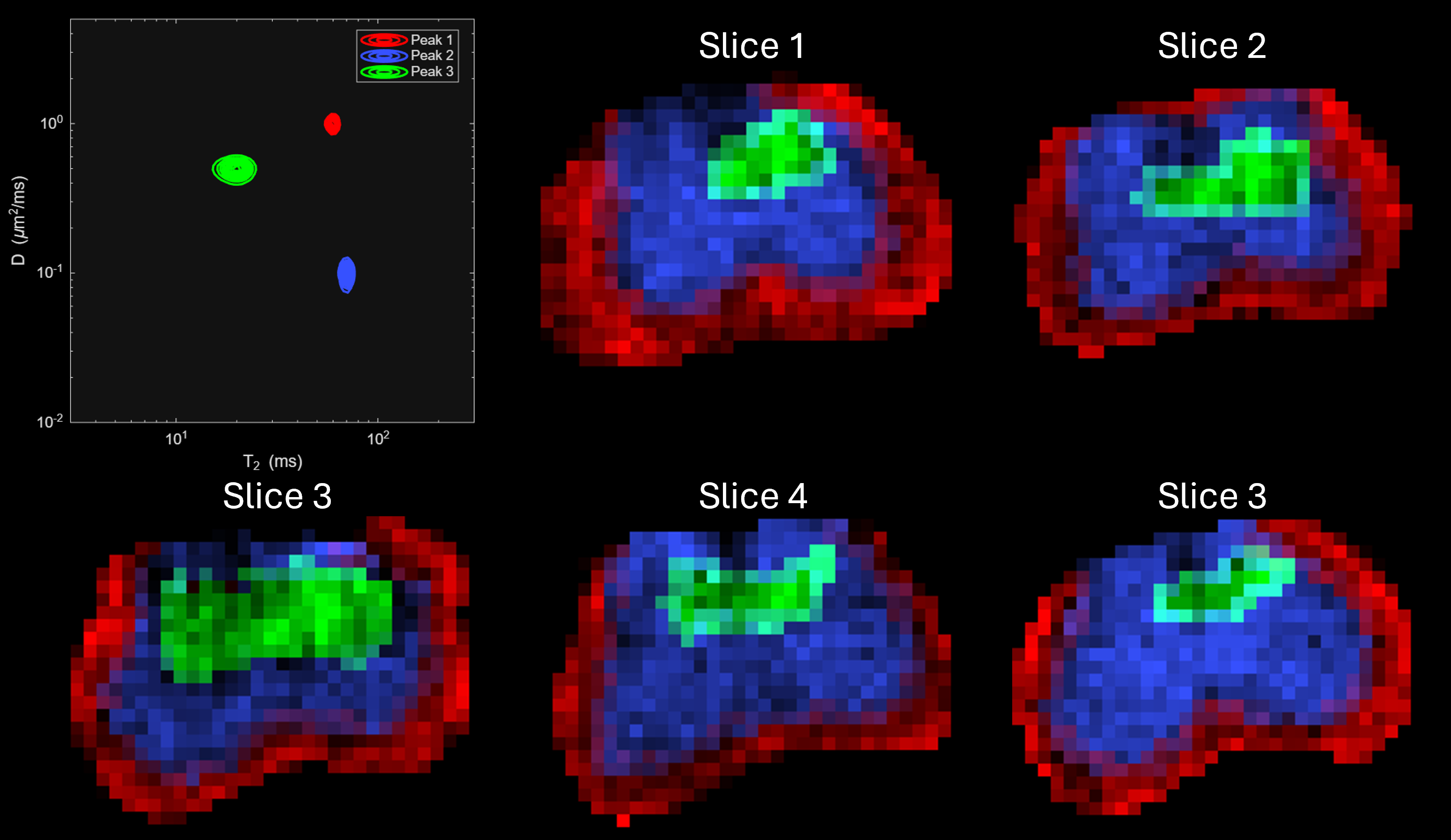
Ground-truth structure of the phantom.
Phantom data can be generated using the following command:
create_phantom.sh -acqfile acqfile_name -spectfile spectfile_name -multislice tf -outfolder outfolder_name
create_phantom('acqfile','___','spectfile','___','outfolder','___','multislice','___')
8.1. Optional Inputs
acqfile:
Filename of the
.txtfile storing combinations of(b, TE)values where MR data will be sampled.bis in ms/µm² andTEis in ms.If omitted, defaults to a built-in 7×7 grid:
b = [0 200 500 1000 1500 2500 5000]*1e-3(\(ms/ \mu m^2\)) andTE = [99 120 160 200 250 300 400](in \(ms\)).The code infers data type and spectrum information automatically:
only TE changes → 1D T2 phantom
only b changes → 1D Diffusion phantom
both change → D–T2 phantom
Create a text file (e.g.
acq_params.txt):b_1,TE_1 b_2,TE_2 ... b_n,TE_n
For 1D Diffusion data, keep
TEconstant:b_1,TE_1 b_2,TE_1 ... b_n,TE_1
For 1D T2-Relaxation data, keep
bconstant:b_1,TE_1 b_1,TE_2 ... b_1,TE_m
Notes:
This Phantom generation tool is very sensitive to this format. If given in other format, the tool wont work.
spectfile:
Filename of the
.txtfile storing information to create spectral sampling points alongDandT2dimensions. Used to build the dictionary matrix for spectrum estimation.If both
acqfileandspectfileare not provided, thespectfiledefaults to the example given on the below.If
acqfileis provided butspectfileis not omitted, then it automatically detects the datatype and uses the default values as given below. While for both the 1D and 2D spectrum, the end points and the spacing type of the spectral grid remains the same, for 1D datatype, it uses 300 spectral grid points by dafult.
Create a text file (e.g.
spect_params.txt):D_min=0.01; D_max=3; D_spacing='log'; D_num=70; T2_min=3; T2_max=300; T2_spacing='log'; T2_num=70;
Notes:
The above example also comprises of the default values in the
spectfile.This Phantom generation tool is very sensitive to the variable names used in the above example. If any other names are used, the tool won’t work.
D_min,D_maxshould be in \(ms/ \mu m^2\);T2_min,T2_maxin \(ms\).D_spacingandT2_spacingcan only take'linear'or'log'.For 1D phantoms, include only relevant parameters (either
DorT2).
multislice:
Logical flag:
1→ Generate five 2D slices.0→ Generate single 2D slice.Default:
0.Example (command line):
create_phantom.sh -multislice 1
outfolder:
Folder name where the output files will be stored. Default:
Phantom(created in the current directory).Example:
create_phantom.sh -outfolder "Phantom_Test"
Warning
The user is recommended to provide an input corresponding to the outfolder.
Not providing a folder name will rewrite the contents of the default folder named Phantom.
8.2. Outputs
The following files are saved in the folder name specified by outfolder:
Phantom_data.mat:
Stores the information related to 2D/3D spatial diffusion-relaxation MR data. Same structure as here (input to spectrum estimation tool).
Phantom_spectrum_info.mat:
Stores information related to the spectrum. Structure follows format here.
Phantom_mask.mat:
Spatial mask file for the MR data. Structure follows format here.
Phantom_mask_beta_calc.mat:
Spatial mask file for (L)ADMM parameter
betacalculation. Structure follows format here.