7.1. Plotting spectroscopic images
Spectroscopic images can be visualized with the following command:
plot_spect_im.sh -spect_imfile spect_imfile_name -imgfile imgfile_name -outprefix outprefix_name
plot_spect_im('spect_imfile','___','imgfile','___','outprefix','___')
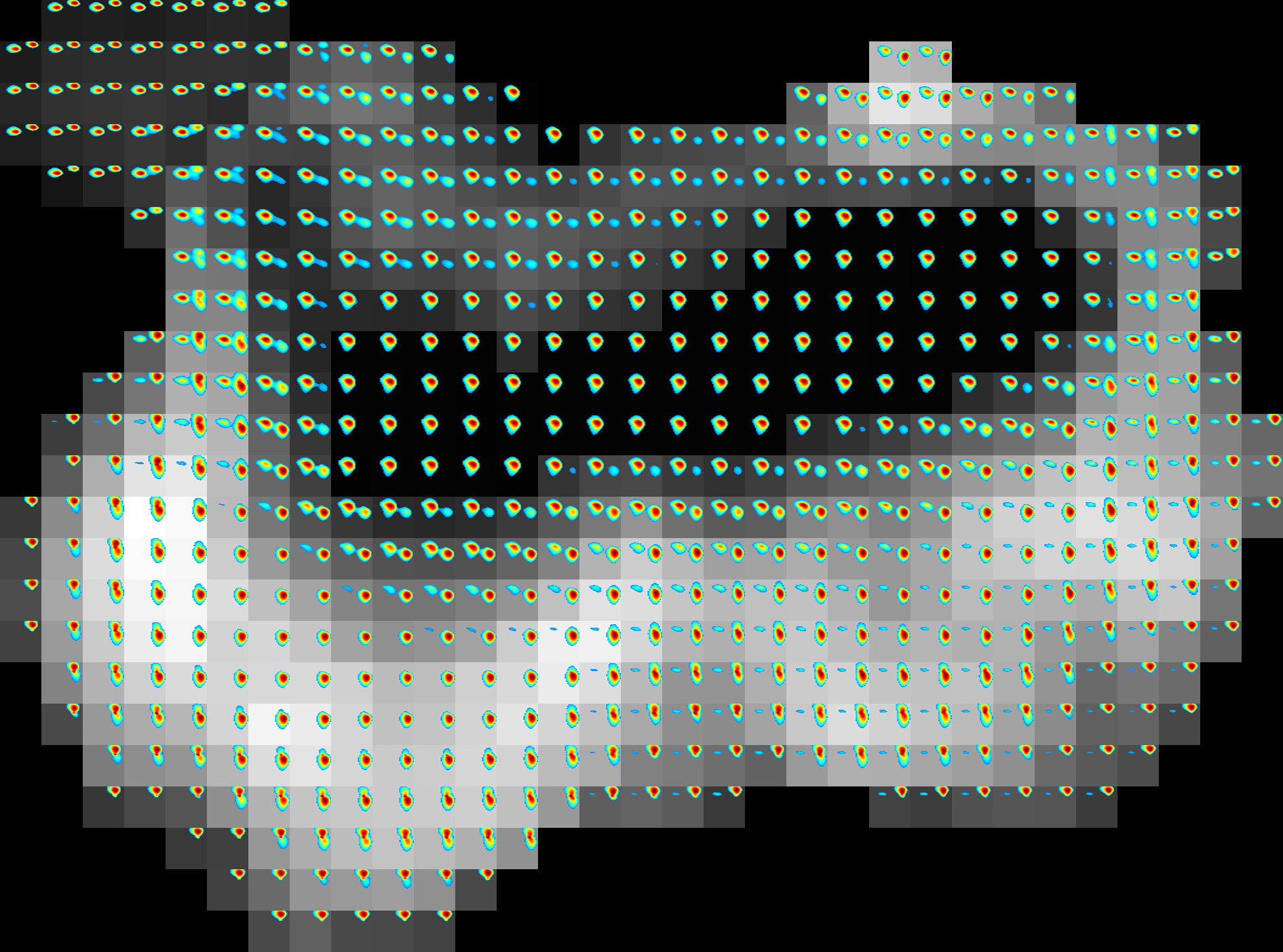
Example of 2D spectroscopic image |
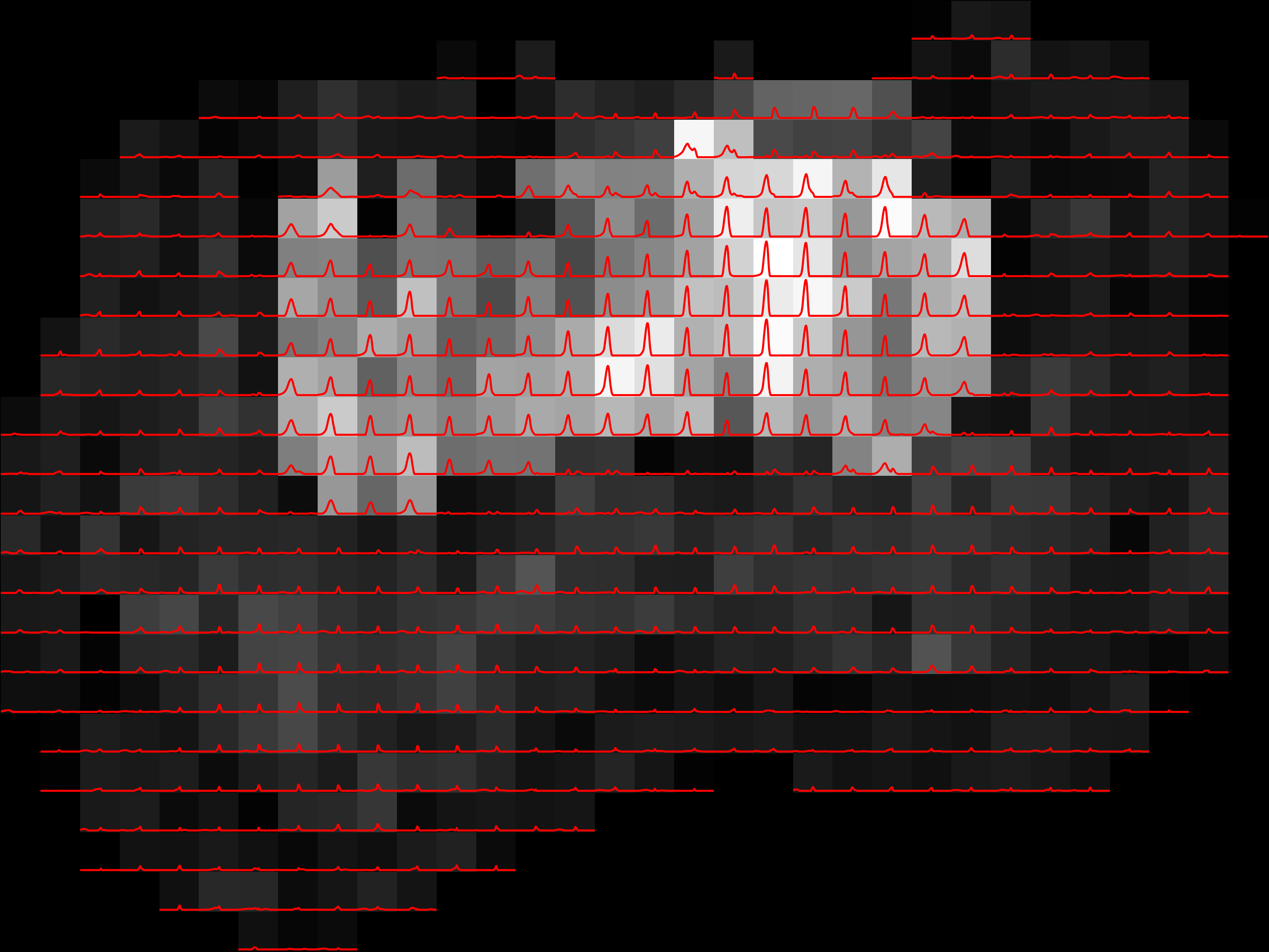
Example of 1D spectroscopic image |
If the spatial volume is 3D then, then the command will ouotput spectroscopic images for each slice. This tool does not currently support more than 2 spectral dimensions. more than 2.
When using optional arguments, the following syntax should be used:
plot_spect_im.sh -spect_imfile spect_imfile_name -imgfile imgfile_name -outprefix outprefix_name -spatmaskfile spatmaskfile_name -ax_scale ax_scale_name -ax_lims ax_lims_values -color color_name -enc_idx enc_idx_val -normalise tf -linewidth linewidth_val -threshold threshold_val -file_types file_types_name
plot_spect_im('spect_imfile','___','imgfile','___','outprefix','___','spatmaskfile','___','ax_scale','___','ax_lims','___','color','___','enc_idx','___','normalise',''___,'linewidth','___','threshold','___','file_types','___')
7.1.1. Required Inputs
spect_imfile::
The spectroscopic image to be visualized. The spectroscopic image must be in the
spectImagefileformat (e.g., as generated through DRSuite’s spectroscopic image estimation tool) as described here.imgfile:
A corresponding set of multicontrast MRI images to use as the background for visualization. This should be given in the
imgfileformat as described here, with the same spatial dimensions as the spectroscopic image, and will often be the same file used as input to spectroscopic image estimation.idx:
An integer specifying which image from
imgfileshould be used as the background.spatmaskfile:
A binary spatial mask to use during visualization. This must be in the
spatmaskfileformat as described here, with the same spatial dimensions as the spectroscopic image, and will often be the same file that was used as input to the spectroscopic image estimation tool.outputprefix:
A string specifying the filename prefix to use for the saved output images. For 3D spatial images, each slice is saved separately using this prefix plus a slice index.
7.1.2. Optional Inputs
ax_scale:
A single string to control scaling of the axes denoting spectral sampling points. Available only for 1D spectrum. Accepts
'linear'or'log'only. If no input is provided, defaults to the spectral axes spacing fromaxes(1).spacingin spectroscopic image file.ax_lims:
Axis limits for the overlayed spectra. For 1D spectra use
[xmin xmax](defaults to the first and last samples fromaxes(1).sample). For 2D spectra use[ymin ymax xmin xmax](defaults fromaxes(1).sampleandaxes(2).sample). If length of this vector is other than 2 or 4 it shows an error.color:
Used to define Line color for 1D spectra or colormap for 2D spectra.
1D spectra: accepts a MATLAB color char/keyword (e.g.,
'r','red'). Recognized color keywords for 1D spectra:'r','g','b','c','m','y','k','w','red','green','blue','cyan','magenta','yellow','black','white'.2D spectra: Accepts a built-in colormap name (e.g.,
'jet'). Recognized colormaps for 2D spectra:'parula','jet','hsv','hot','cool','spring','summer','autumn','winter','gray','bone','copper','pink','lines','colorcube','prism','flag','white','turbo'.
linewidth:
Line width for 1D line plots and 2D contour plots (default: 1.2).
threshold:
A fraction in \([0,1]\) that controls the masking of the voxelwise 2D spectral overlays by the MR data. Higher the value, higher the masking of the 2D spectra by the MR data volume. Not used for plotting spectroscopic image of 1D spectra.
figure_types:
A string specifying the output image format (which will also be used as the filename suffix). Available options (not case sensitive):
fig,eps,pdf,bmp,jpg,png,tif,tiff,svg.
7.1.3. Output
Image files are saved using the specified filename prefix and output format.